Home
Fig. 1 A screenshot of the home page
Home page
The function of the Home page is to introduce msRepDB, mainly including the application fields of msRepDB, the research and development basis, and the main advantages compared with the existing libraries.
Operation example:
1) Click the "Home" option in the navigation bar;
[Server response]: The server will display the home page in the center of the screen (Fig. 1).
2) Slide the mouse wheel;
[Server response]: The server will display the rest of the page in the center of the screen.
Search and Download
Fig. 2 A screenshot of the Search and Download page (1)
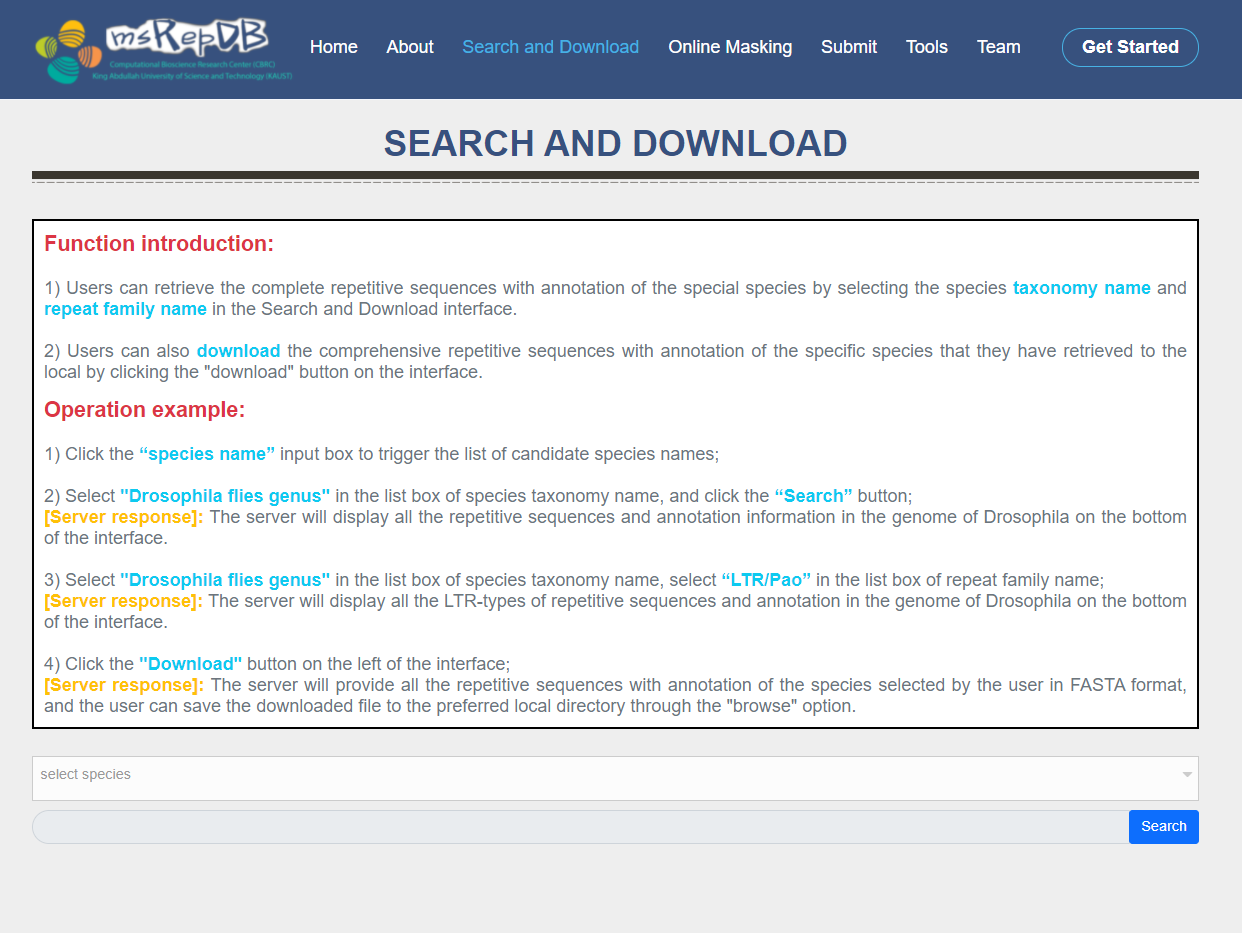
Search and Download page
The functions of the Search and Download page are as follows:
1) By selecting the species taxonomy name and repeat family name in the Search and Download interface, the Users can retrieve the complete repetitive sequences with annotation of the special species (Fig. 2). 2) By clicking the "download" button on the interface, the users can also download the comprehensive repetitive sequences with annotation of the specific species that they have retrieved to the local disk (Fig. 3).
Operation example:
1) Click the "select species" input box to trigger the list of candidate species names;
Fig. 3 A screenshot of the Search and Download page (2)
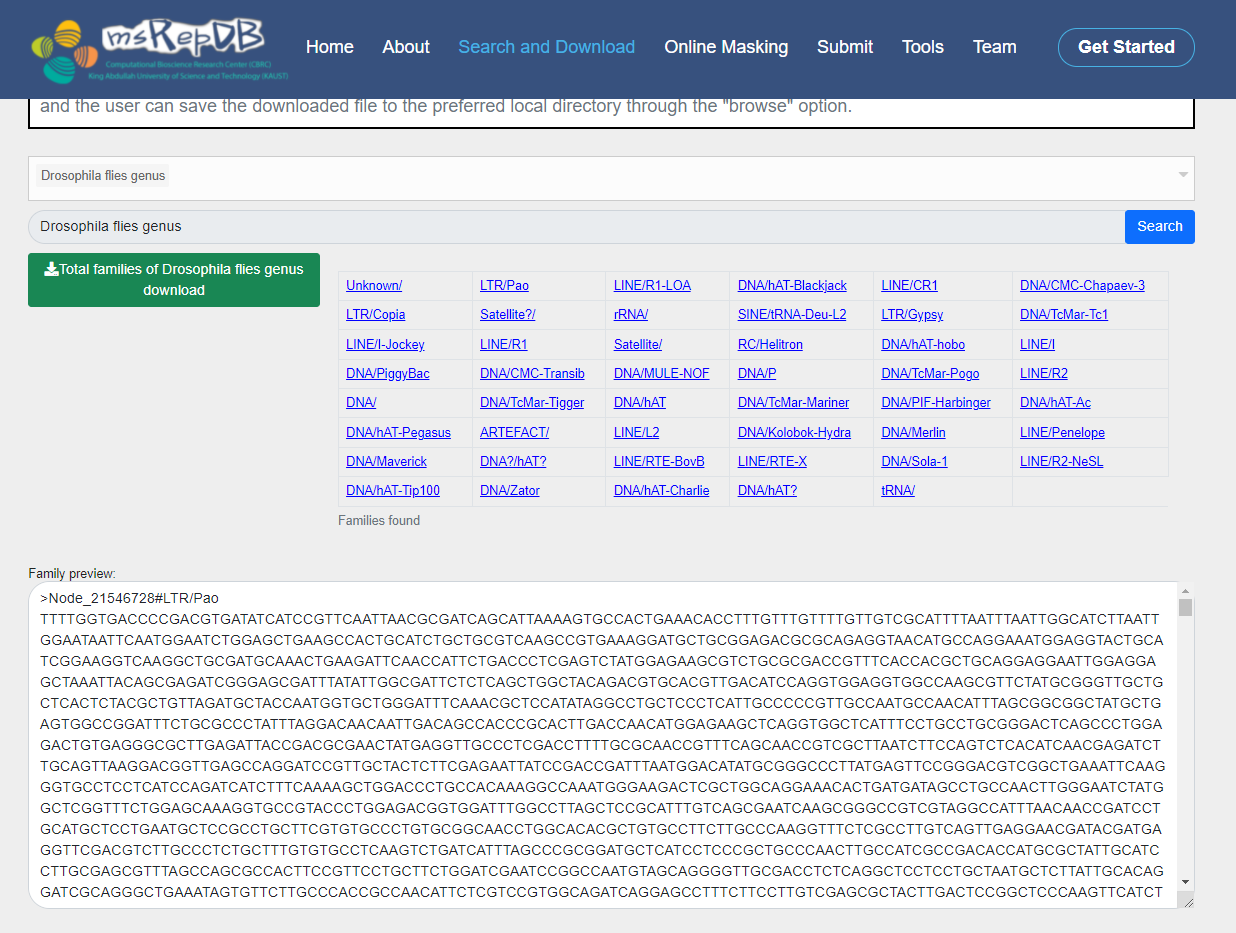
2) Select "Drosophila files genus" in the list box of species taxonomy name, and click the “Search” button;
[Server response]: The server will display all the repetitive sequences and annotation in the genome of Drosophila on the bottom of the interface.
3) Select "Drosophila files genus" in the list box of species taxonomy name, select “LTR/Pao” in the list box of repeat family name, and click the “Search” button;
[Server response]: The server will display all the LTR/Pao-types of repetitive sequences and annotation in the genome of Drosophila on the bottom of the interface.
4) Click the "Total families of Drosophila files genus download" button on the left of the interface;
[Server response]: The server will provide all the repetitive sequences with annotation (LTR/Pao) of the species selected (Drosophila files genus) by the user in FASTA format, and the user can save the downloaded file to the preferred local directory through the "browse" option.
Online Masking
Fig. 4 A screenshot of the Online Masking page (1)
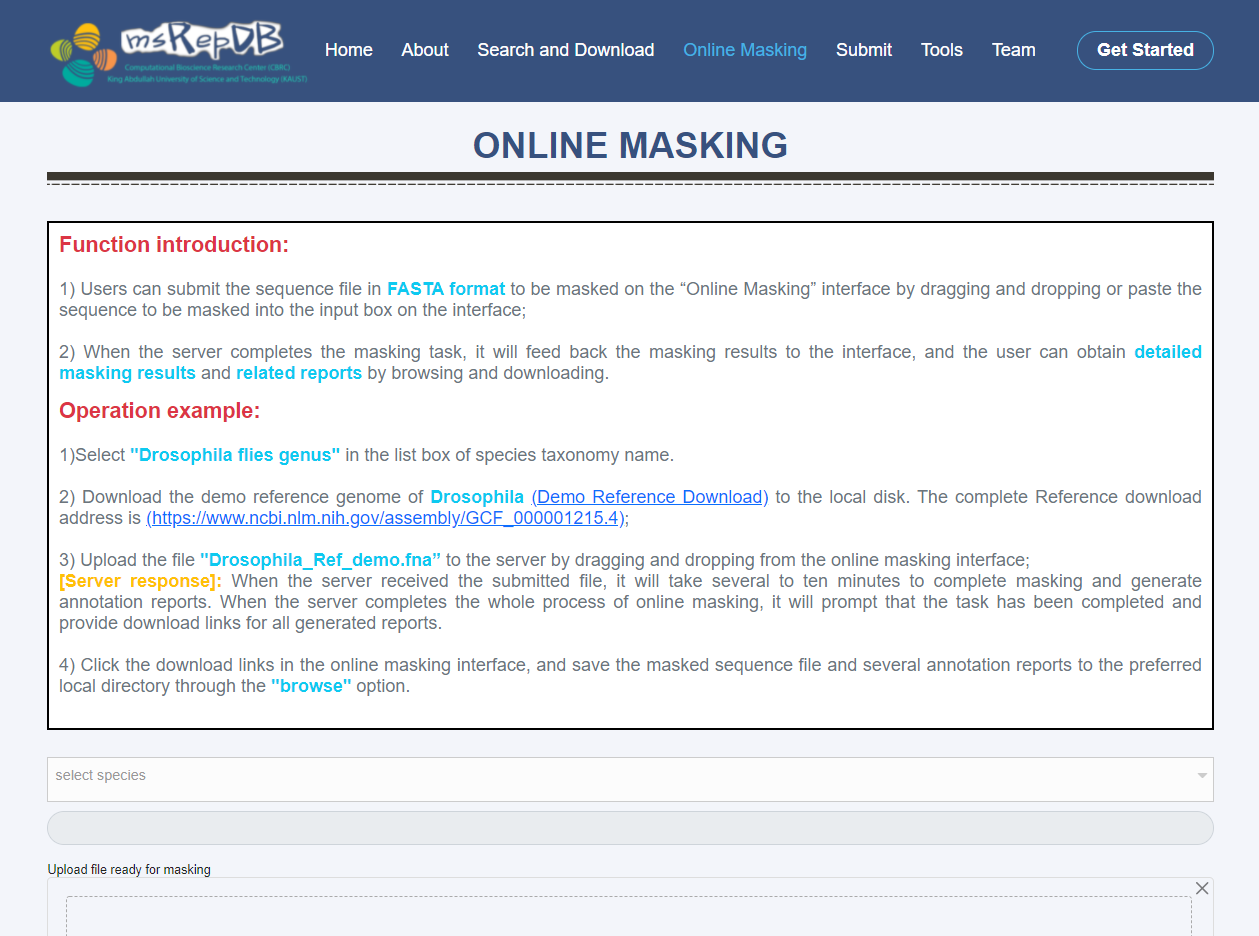
Online Masking page
The functions of the Online Masking are as follows:
1) By dragging and dropping or paste the sequence to be masked into the input box on the interface, the users can submit the sequence file in FASTA format to be masked on the “Online Masking” interface (Fig. 5);
2) When the server completes the masking task, it will feed back the masking results to the interface, and the user can obtain detailed masking results and related reports by browsing and downloading (Fig. 6).
Fig. 5 A screenshot of the Online Masking page (2)
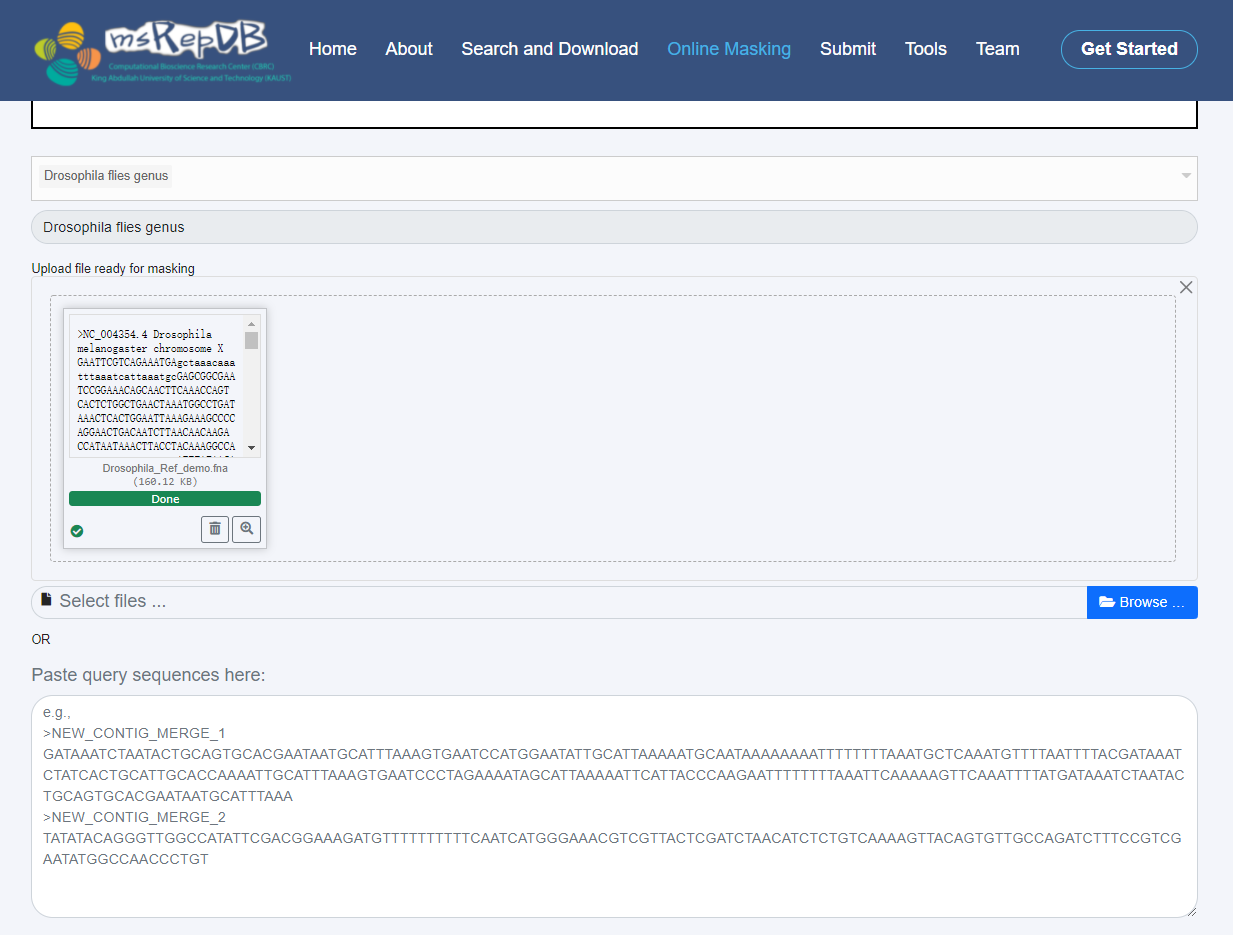
Operation example:
1) Select "Drosophila files genus" in the list box of species taxonomy name;
2) Download the demo reference genome of Drosophlia (Demo_Refence_Download) to the local disk. The complete Reference download address is https://www.ncbi.nlm.nih.gov/ assembly/GCF_000001215.4;
3) Upload the file "Drosophila_Ref_demo.fna" to the server by dragging and dropping from the online masking interface;
Fig. 6 A screenshot of the Online Masking page (3)
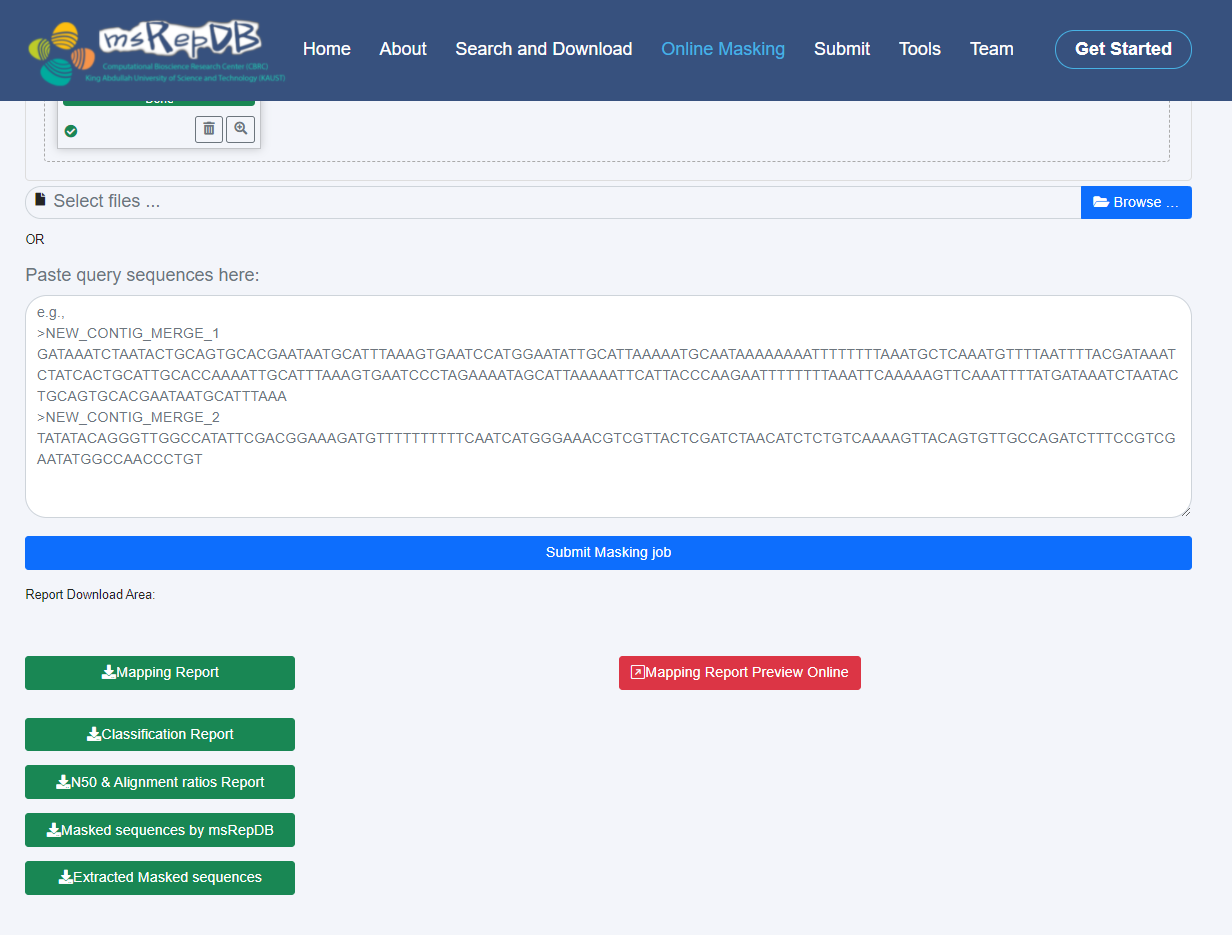
4) Click the "Submit Masking job" button;
[Server response]: When the server received the submitted file, it will take several to ten minutes to complete masking and generate annotation reports. When the server completes the whole process of online masking, it will prompt that the task has been completed and provide download links for all generated reports.
5) Click the download links (such as "Mapping Report","Classification Report","N50 & Alignment ratios Report","Masked sequences by msRepDB" and "Extracted Masked Sequences") in the online masking interface, and save the masked sequence file and several annotation reports to the preferred local directory through the "browse" option;
[Server response]: The server will provide download links for the masked sequence files and all generated reports.
Submit
Fig. 7 A screenshot of the Submit page (1)
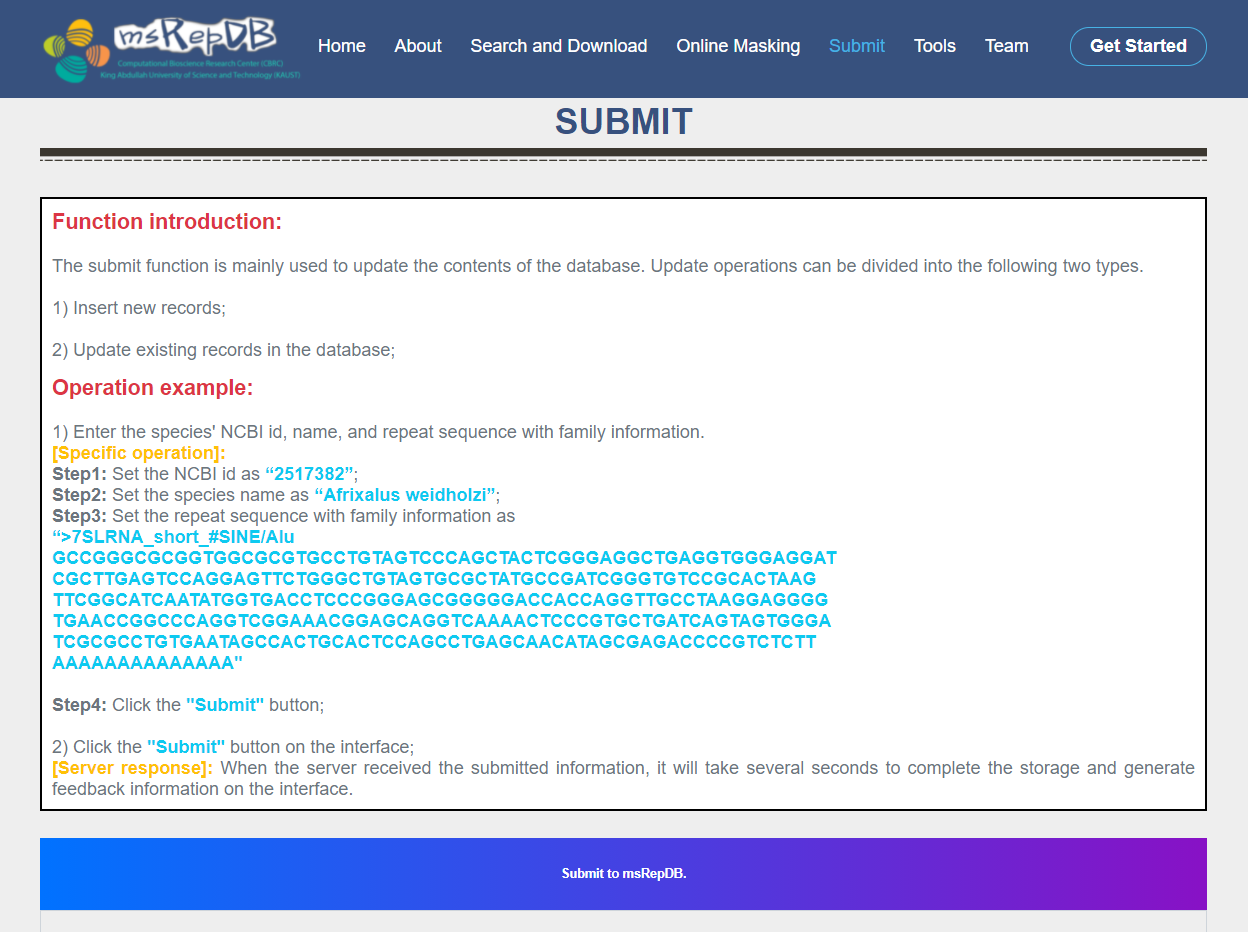
Submit page
The submit function is mainly used to update the contents of the database (Fig. 7). Update operations can be divided into the following two types:
1) Insert new records. 2) Update existing records in the database.
Operation example:
1) Enter the species' NCBI id, name, and repeat sequence with family information (Fig. 8);
[Specific operation]:
Step1: Set the NCBI id as “2517382”;
Step2: Set the species name as “Afrixalus weidholzi”;
Fig. 8 A screenshot of the Submit page (2)
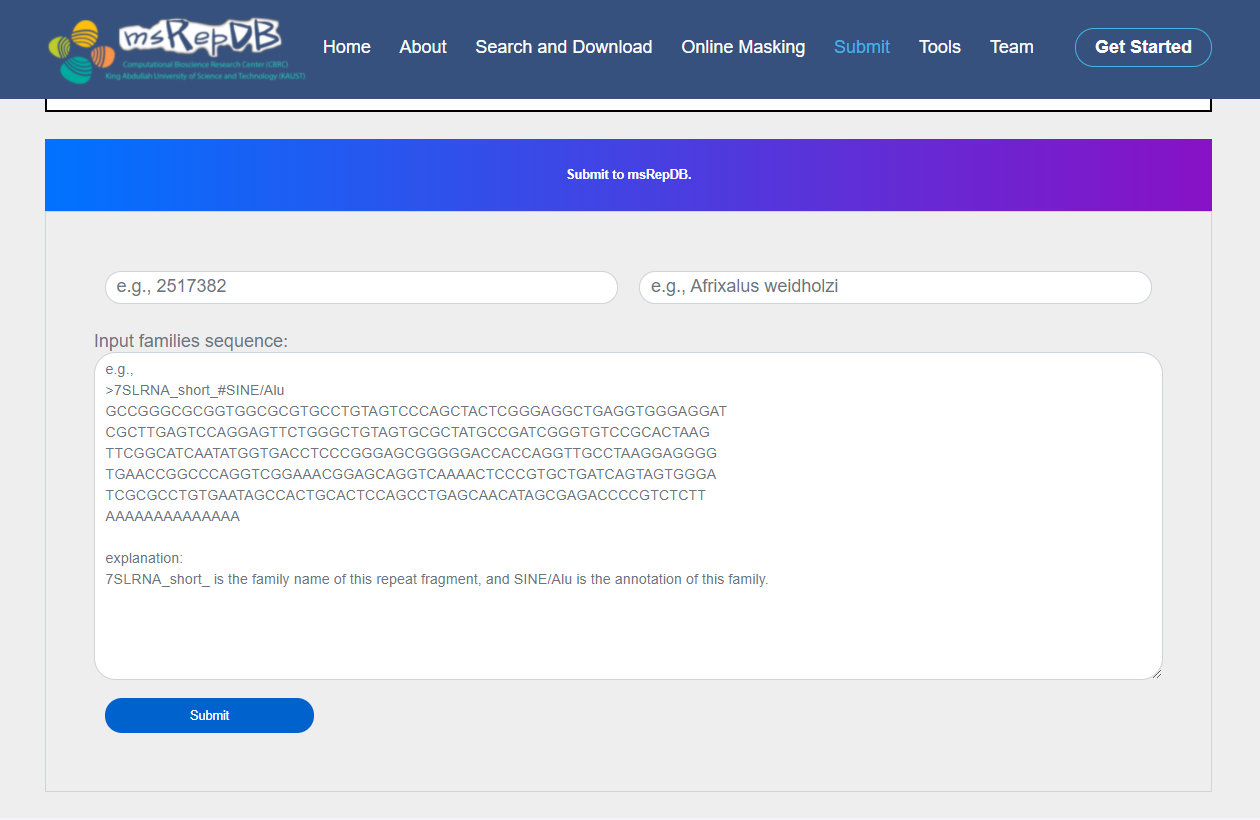
Step3: Set the repeat sequence with family information as “>7SLRNA_short_#SINE/Alu GCCGGGCGCGGTGGCGCG TGCCTGTAGTCCCAGCTA CTCGGGAGGCTGAGGTGG GAGGATCGCTTGAGTCCA GGAGTTCTGGGCTGTAGT GCGCTATGCCGATCGGGT GTCCGCACTAAGTTCGGC ATCAATATGGTGACCTCC CGGGAGCGGGGGACCACC AGGTTGCCTAAGGAGGGG TGAACCGGCCCAGGTCGG AAACGGAGCAGGTCAAAA CTCCCGTGCTGATCAGTA GTGGGATCGCGCCTGTGA ATAGCCACTGCACTCCAG CCTGAGCAACATAGCGAG ACCCCGTCTCTTAAAAAA AAAAAAAA”;
2) Click the "Submit" button on the interface;
[Server response]: When the server received the submitted information, it will take several seconds to complete the storage and generate feedback information on the interface.
Tools
Fig. 9 A screenshot of the Tools page
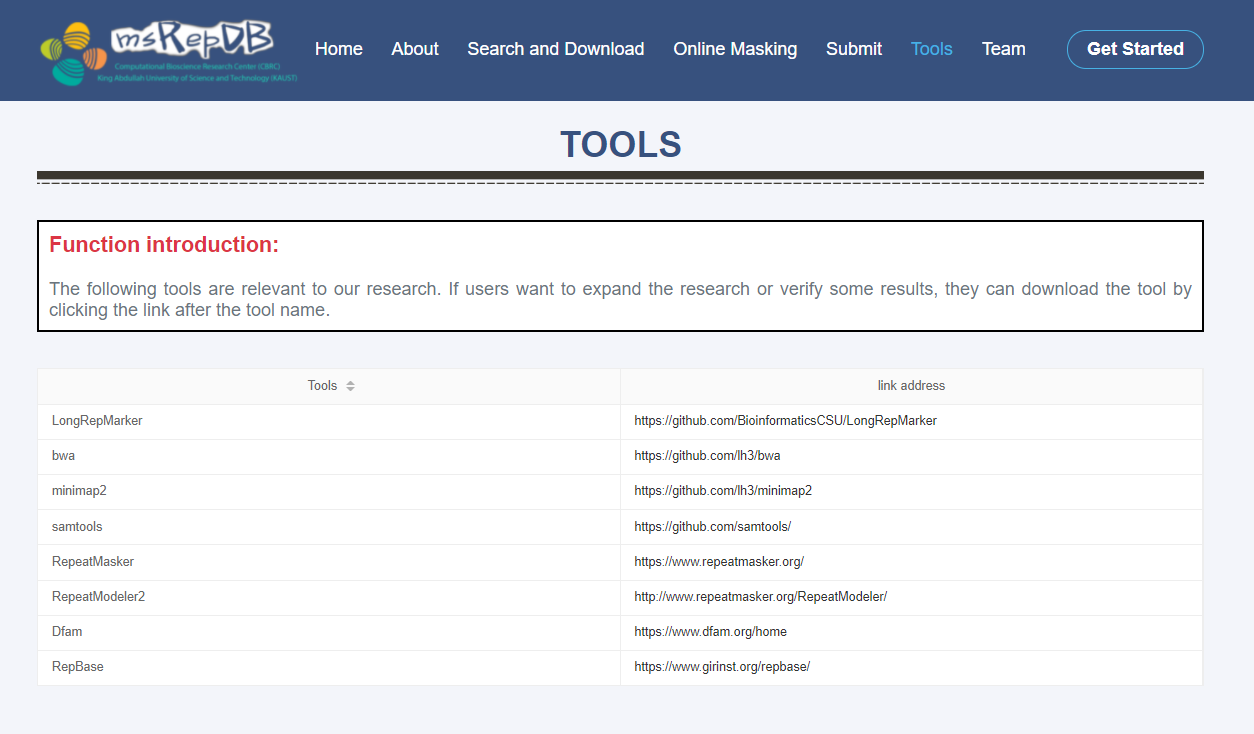
Tools page
The following tools are relevant to our research. If users want to expand the research or verify some results, they can download the required tool by clicking the link after the tool name (Fig. 9).
Team
Fig. 10 A screenshot of the Team introduction page
Team members:
1) Xingyu Liao [First Author], [Project designer] Postdoctoral Fellow, Computational Bioscience Research Center (CBRC), Computer, Electrical and Mathematical Sciences and Engineering Division, King Abdullah University of Science and Technology (KAUST), Thuwal, 23955, Saudi Arabia. E-mail: xingyu.liao@kaust.edu.sa URL: https://cemse.kaust.edu.sa/cbrc/people/person/xingyu-liao
2) Kang Hu [Second Author], [Core developer] Ph.D. Student, Hunan Provincial Key Lab on Bioinformatics, School of Computer Science and Engineering, Central South University, Changsha 410083, P.R. China. E-mail: kanghu@csu.edu.cn
3) Adil Salhi [Third Author], [Project operation and maintenance personnel] Research Engineer, Structural and Functional Bioinformatics Group, Postdoctoral Fellows, Knowledge Mining Lab, King Abdullah University of Science and Technology (KAUST), Thuwal, 23955, Saudi Arabia. E-mail: adil.salhi@kaust.edu.sa URL: https://cemse.kaust.edu.sa/cbrc/people/person/adil-salhi
4) Xin Gao [Corresponding Author], [Project manager] Professor, Computer Science, Principal Investigator, Structural and Functional Bioinformatics Group, Acting Associate Director, Computational Bioscience Research Center (CBRC), King Abdullah University of Science and Technology (KAUST), Thuwal, 23955, Saudi Arabia. E-mail: xin.gao@kaust.edu.sa URL: https://cemse.kaust.edu.sa/cbrc/people/person/xin-gao